Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the virus behind the worldwide outbreak of COVID-19 disease. One of the key biological characteristics of SARS-CoV-2, as well as several other viruses, is the presence of spike proteins that allow these viruses to penetrate host cells and cause infection.
Image Credit: MattLphotography/Shutterstock.com
An overview of the coronavirus structure
Coronaviruses (CoVs) like the Middle East respiratory syndrome (MERS)-CoV, which had infected almost 2,500 individuals by the end of 2019, as well as the novel severe acute respiratory syndrome (SARS)-CoV-2, are enveloped and spherical viruses that typically measure between 80 and 120 nanometers (nm) in size.
The RNA genome of coronaviruses, which, at a median length of 29 kb is the longest among all RNA viruses, is comprised of six to ten open reading frames (ORFs) that are responsible for encoding both the replicase and structural proteins for the virus.
Each of the components of the viral genome is packaged into a helical nucleocapsid that is surrounded by a lipid bilayer.
The viral envelope of coronaviruses is typically made up of three proteins that include the membrane protein (M), the envelope protein (E), and the spike protein (S).
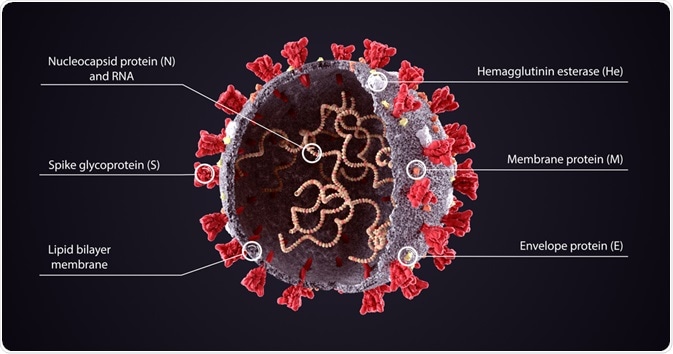
Image Credit: Orpheus FX/Shutterstock.com
Spike protein functions
The S protein is a highly glycosylated and large type I transmembrane fusion protein that is made up of 1,160 to 1,400 amino acids, depending upon the type of virus.
As compared to the M and E proteins that are primarily involved in virus assembly, the S protein plays a crucial role in penetrating host cells and initiating infection.
Notably, the presence of S proteins on the coronaviruses is what gives rise to the spike-shaped protrusions found on their surface.
S proteins of coronaviruses can be divided into two important functional subunits, which include the N-terminal S1 subunit, which forms the globular head of the S protein, and the C-terminal S2 region that forms the stalk of the protein and is directly embedded into the viral envelope.
Upon interaction with a potential host cell, the S1 subunit will recognize and bind to receptors on the host cell, whereas the S2 subunit, which is the most conserved component of the S protein, will be responsible for fusing the envelope of the virus with the host cell membrane.
How S protein allows coronaviruses to enter cells
Once the S1 subunit binds to host cell receptors, two major conformational changes must occur for the S2 subunit to complete the fusion of the virus to the cell membrane. The two components of the S2 subunit that are involved in the coronavirus fusion include heptad repeat (HR) regions one and two, otherwise referred to as HR1 and HR2.
The first conformation otherwise referred to as pre-hairpin, involves the transformation of an unstructured linker within the S2 subunit to become helical. The second conformational change to occur involves the inversion of this subunit’s C-helix to the coil, resulting in the formation of a six-helix bundle.
Once these conformations are completed, the fusion peptide is anchored to the membrane of the host cell to allow the virus to move closer towards the cell membrane and eventually deliver the nucleocapsid to the target cell.
Targeting viruses via the S protein
Without the S protein, viruses like the novel SARS-CoV-2 would not be able to interact with the cells of potential hosts like animals and humans to cause infection. As a result, the S protein represents an ideal target for vaccine and antiviral research endeavors.
In addition to its role in penetrating cells, the S protein of viruses, particularly the SARS-CoV-2 virus, is a major inducer of neutralizing antibodies (NAbs). NAbs are protective antibodies that are naturally produced by our humoral immune system.
To elicit their antiviral activity, NAbs bind to the surface epitopes of viral particles to prevent their entry into a host cell. Some of the NAbs that have been identified against SARS-CoV include 80R, CR3014, and CR3022, all of which specifically target the S1 domain.
The sensitivity of the S protein of SARS-CoV-2 to these NAbs has led many researchers to be interested in developing standardized agents that can block the binding and fusion of the S protein of SARS-CoV-2 to host cells.
Although one recent study has found that the neutralization capability of CR3022 against the SARS-CoV-2 region binding domain (RBD) within the S1 subunit is uncertain, another study on both Ebola and SARS viruses has found that a cocktail of NAbs has stronger neutralization capabilities as compared to when these NAbs are given alone.
This information, therefore, leads researchers to believe that a combination of several potent NAbs has the potential to target SARS-CoV-2 and increase its sensitivity to neutralization.
References
- Belouzard, S., Millet, J. K., Licitra, B. N., & Whittaker, G. R. (2012). Mechanisms of Coronacirus Cell Entry Mediated by the Viral Spike Protein. Viruses 4(6); 1011-1033. doi:10.3390/v4061011.
- Wang, Q., Wong, G., Lu, G., Yan, J., & Gao, G. F. (2016). MERS-CoV spike protein: Targets for vaccines and therapeutics. Antiviral Research 133; 165-177. doi:10.1016/j.antiviral.2016.07.015.
- Zhou, G., & Zhao, Q. (2020). Perspectives on therapeutic neutralizing antibodies against the Novel Coronavirus SARS-CoV-2. International Journal of Biological Sciences 16(10); 1718-1723. doi:10.7150/ijbs.45123.
Further Reading